Open student projects
Explore our available student projects!
Visual Language Models for Multimodal Data Fusion in Low Back Pain Research
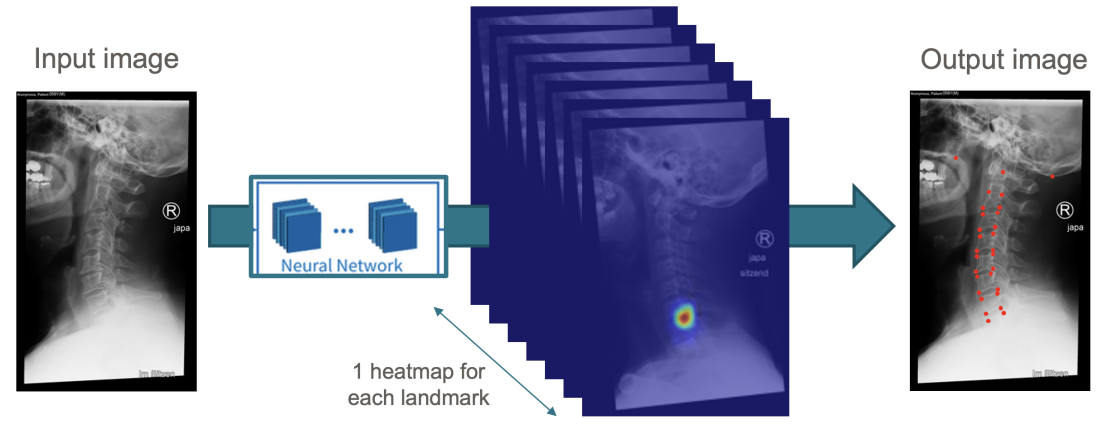
Supervisor
Maria Monzon
Aim
To develop and evaluate a visual language model for multimodal data fusion in low back pain research, with primary focus on treatment outcome-based patient stratification and secondary emphasis on automated MRI scan caption generation.
- Conduct a comprehensive literature review on state-of-the-art visual language models [3], multimodal data fusion techniques in medical imaging, and current applications of AI in low back pain research
- Understand the structure of raw data and build a multimodal dataset, including MRI scans of the lumbar spine, clinical patient-reported outcomes and demographic information. Conduct basic exploratory data analysis to understand the relationship between MRI and the presence/severity of low back pain in participants
- Benchmark a visual language model architecture capable of processing and integrating multiple data modalities [1, 2, 3]
- Train and optimize the model using the collected multimodal dataset, employing techniques such as transfer learning, fine-tuning [4] or transduction [5] to adapt existing VLM architectures [6,7] to the LBP domain
- Compare the performance of the multimodal approach to (unimodal) baselines [8] and existing clinical prediction models, ensuring
robustness and reliability across diverse patient populations - Report findings, along with structured code, in a GitLab repository
- Prepare visualizations and analyses that could potentially lead to contributions of a research paper
References
[1] Mohsen, Farida et al. “Artificial intelligence-based methods for fusion of electronic health records and imaging data.” Scientific Reports 12
(2022)
[2] Huang, SC., Pareek, A., Seyyedi, S. et al. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. npj Digit. Med. 3, 136 (2020). https://doi.org/10.1038/s41746-020-00341-z
[3] Xu, Li et al. “Multi-modal Pre-training for Medical Vision-language Understanding and Generation: An Empirical Study with A New Benchmark.” ArXiv abs/2306.06494 (2023):
[4] Lu, Yuzhe et al. “Effectively Fine-tune to Improve Large Multimodal Models for Radiology Report Generation.” ArXiv abs/2312.01504 (2023)
[5] Zanella, Maxime et al. “Boosting Vision-Language Models with Transduction.” ArXiv abs/2406.01837 (2024)
[6] Moor, Michael et al. “Med-Flamingo: a Multimodal Medical Few-shot Learner.” ArXiv abs/2307.15189 (2023)
[7] Zhang, Sheng et al. “BiomedCLIP: a multimodal biomedical foundation model pretrained from fifteen million scientific image-text pairs.” (2023).
[8] Windsor, Rhydian et al. “SpineNetV2: Automated Detection, Labelling and Radiological Grading Of Clinical MR Scans.” ArXivabs/2205.01683 (2022)
- Contribution to a publishable research paper
- Development and application of software engineering skills, including package management, testing practices, and version control
- Our goal is to provide you with an introduction to applied data science (programming, analysis, and visualization) to solve biomedical questions
- As part of our research group, you will take part in our weekly lab meetings, during which we provide updates on current projects and review the latest biomedical and data science literature
- Student in Health Sciences and technology, Biomedical Engineering, Neuroscience, Human Movement Sciences, Biology, Data Science
Computer Science, or related fields of study - Strong background in machine learning, deep learning, and computer vision
- Proficient in Python programming and experience with deep learning frameworks (e.g., PyTorch, MONAI)
- Interest in medical imaging and healthcare applications of AI
- Disciplined, reliable, friendly, and able to work in a team
- Duration: 6 months
- Start Date: March - or as soon as possible
- Full time preferred but part time also possible
- Fotmat: All projects will be conducted at the BMDS lab. If permitted, the research is conducted in the lab at Schulthess clinic (Lengghalde 2, 8008 Zürich). Hybrid setup work with two to four days at the office and the rest from home
- Contact: Please submit a short CV and a record of your studies to: Maria Monzon (]). You can also use this address to ask for further information and in case of questions. We are looking forward to receiving your application.
Developing a Package for Standardized Imputation of a Large Spinal Cord Injury Dataset

Supervisor
Hugo Madge Leon
Aim
To develop a Python package for researchers to generate a reproducible and optimally imputed SCI dataset for recovery prediction models based on the EMSCI data.
The significant challenge of missing data in the context of rare diseases, particularly spinal cord injury (SCI), restricts the early prediction of patients' functional outcomes. Accurate forecasting of recovery trajectories is vital not only for guiding clinical care, but also for managing patient expectations and guiding clinical trial designs [1,2]. The absence of a large, comprehensive dataset for SCI with clearly documented preprocessing practices, especially in dealing with missing data, significantly impedes the ability to compare recovery prediction methods across studies. Therefore, there is a need for robust and transparent data management strategies that can enhance the reliability and comparability of predictive models for patient outcomes.
Work has already been carried out within our lab to tackle this issue by generating an imputation pipeline for data from the European Multicenter Study About Spinal Cord Injury (EMSCI) [3] dataset. This work imputed neurological assessment data from the EMSCI dataset using two multiple imputation methods: predictive mean matching and random forests. Recovery prediction performance of the imputed data was assessed using four distinct recovery prediction models from the literature for lower extremity motor score, walking ability, bowel and bladder function recovery. This work highlighted the potential of imputation for SCI recovery prediction models, which could be of great use to the research community if made into an easy-to-use tool.
References
[1] Kirshblum, S.C., Priebe, M.M., Ho, C.H., Scelza, W.M., Chiodo, A.E., Wuermser, L.-A., 2007. Spinal Cord Injury Medicine. 3. Rehabilitation Phase After Acute Spinal Cord Injury. Arch. Phys. Med. Rehabil. 88, S62–S70. https://doi.org/10.1016/j.apmr.2006.12.003
[2] Chay W and Kirshblum S. Predicting Outcomes After Spinal Cord Injury. Physical Medicine and Rehabilitation Clinics of North America (2020)
[3] European Multicenter Study about Spinal Cord Injury (EMSCI), https://www.emsci.org/
Anomaly Detection Benchmarking for Low Back Pain Patients MRI
Supervisor
Maria Monzon
Aim
To develop and benchmark machine learning models for anomaly detection in MRI scans of the lumbar spine, focusing on identifying and classifying anomalies associated with low back pain.
Low back pain (LBP) is a prevalent condition that significantly impacts individuals' quality of life and poses a substantial economic burden due to healthcare costs and lost productivity. Magnetic Resonance Imaging (MRI) is a critical tool for diagnosing conditions associated with LBP, such as disc herniations, spinal stenosis, and other spinal pathologies. However, the interpretation of MRI scans is often subjective, leading to variability in diagnoses and treatment plans. This variability can be attributed to the complex nature of spinal anomalies, which are sometimes difficult to detect and classify accurately.
The development of robust anomaly detection models for MRI scans can enhance the diagnostic process by providing consistent and objective assessments of spinal anomalies. Such models can assist radiologists in identifying atypical patterns that may indicate underlying pathologies. The challenge lies in creating models that are sensitive enough to detect subtle anomalies while maintaining specificity to reduce false positives. Benchmarking these models against a standardized dataset can provide insights into their performance and guide further improvements.
- Understand the structure of MRI data, and combine with clinical annotations to build comprehensive datasets for each participant suitable for anomaly detection. Conduct basic exploratory data analysis to understand the relationship between MRI and the presence/severity of low back pain in participants.
- Preprocess and clean the MRI data, standardizing image quality and applying necessary image enhancements.
- Explore generative methods [1] for medical imaging for the creation of synthetic dataset for anomaly detection.
- Develop baseline methods for anomaly detection based on BMAD library [2].
- Evaluate and compare the performance of different models using appropriate metrics.. Analyze areas of strength and weakness for each approach.
- (Optional) Create a demo application to integrate the method developed as an API.
- Report the findings, along with code and documentation, in a Gitlab repository. Prepare visualizations and analyses that could potentially lead to contributions of a research paper.
References
[1] Saha, A., Harowicz, M., Grimm, L., Kim, C., Ghate, S., Walsh, R., & Mazurowski, M.. (2021). Dynamic contrast-enhanced magnetic resonance images of breast cancer patients with tumor locations
[2] Jinan Bao, Hanshi Sun, Hanqiu Deng, Yinsheng He, Zhaoxiang Zhang, & Xingyu Li. (2024). BMAD: Benchmarks for Medical Anomaly Detection.
- Student in Health Sciences and technology, Biomedical Engineering, Neuroscience, Human Movement Sciences, Biology, Data Science, Computer Science, or related fields of study
- Knows essential image processing and/or deep learning and is willing to learn more during the project.
- Good Python programming skills (optional familiar with machine learning frameworks such as PyTorch or Monai)
- Disciplined, reliable, friendly, and able to work in a team